Resources
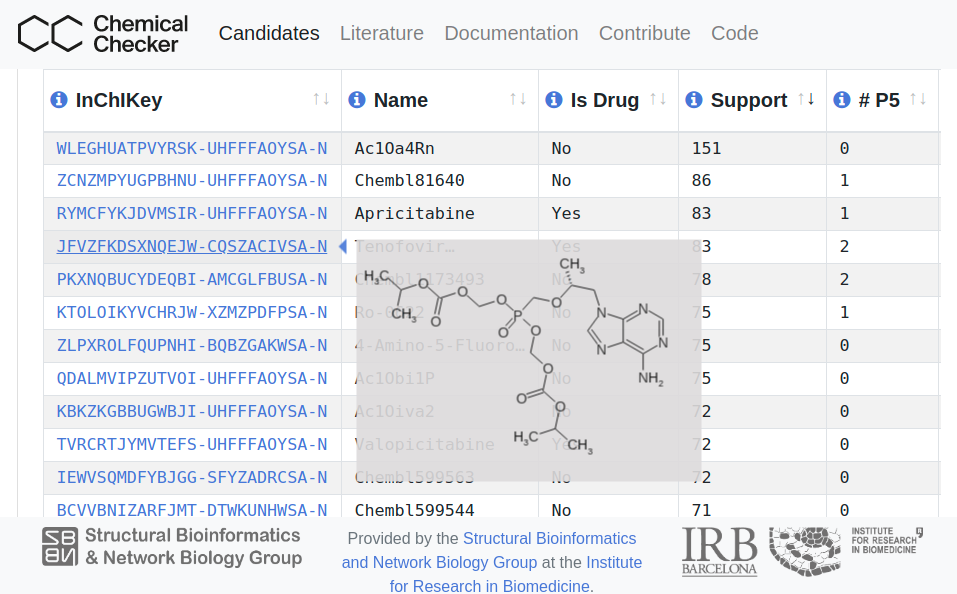
COVID19 Chemical Checker Drug Candidates
Developed by Structural Bioinformatics and Network Biology group, IRB Barcelona in collaboration with the Amazon Search Science and AI group on the NLP tasks.
List of bioactive chemical compounds with potential to be effective against COVID-19. Active collection of suggested drugs from the current COVID-19 literature, with different levels of supporting evidence. Chemical Checker is used to identify small molecules with similar chemical and bioactivity features to the reported drugs in a universe of 800 thousand bioactive compounds.

Covid19 pathway interpretation and analysis (CoV-Hipathia)
Developed by Clinical Bioinformatics Area, FPS
Web tool implementing a mechanistic model of human signalling for the interpretation of the consequences of the combined changes of gene expression levels and/or genomic mutations in the context of signalling pathways. It includes first versions of affected pathways by Covid-19.
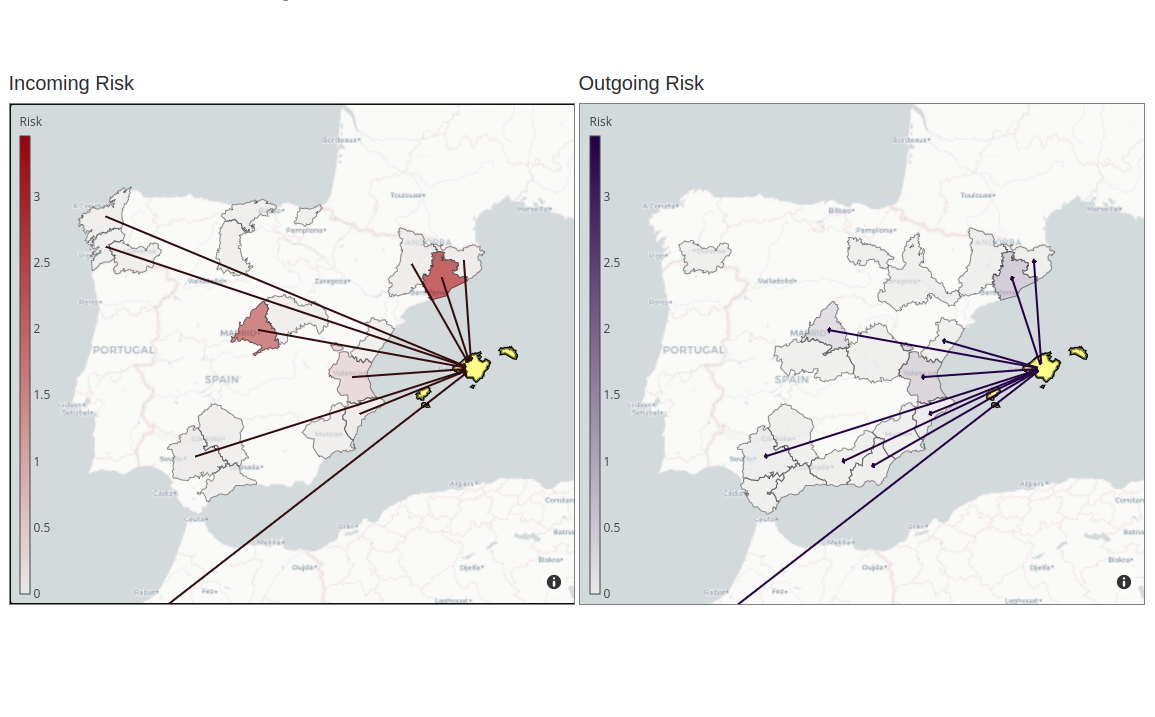
COVID-19 Flow-Maps
Developed by Computational Biology group and INB/ELIXIR-ES coordination team, BSC
System for monitoring COVID-19 outbreaks and mobility-associated risk by integrating health information, population-level mobility patterns into a Geographical Information System.
COVID-19 Viral Beacon
Developed by EGA team, CRG
Tool to find SARS-CoV-2 variability at genomic, amino acid and motif level. It has been developed as a branch of the GA4GH Beacon standard, as a special use case for testing and demonstration of new features in Beacon v2 (and implicitly of Beacon v1).
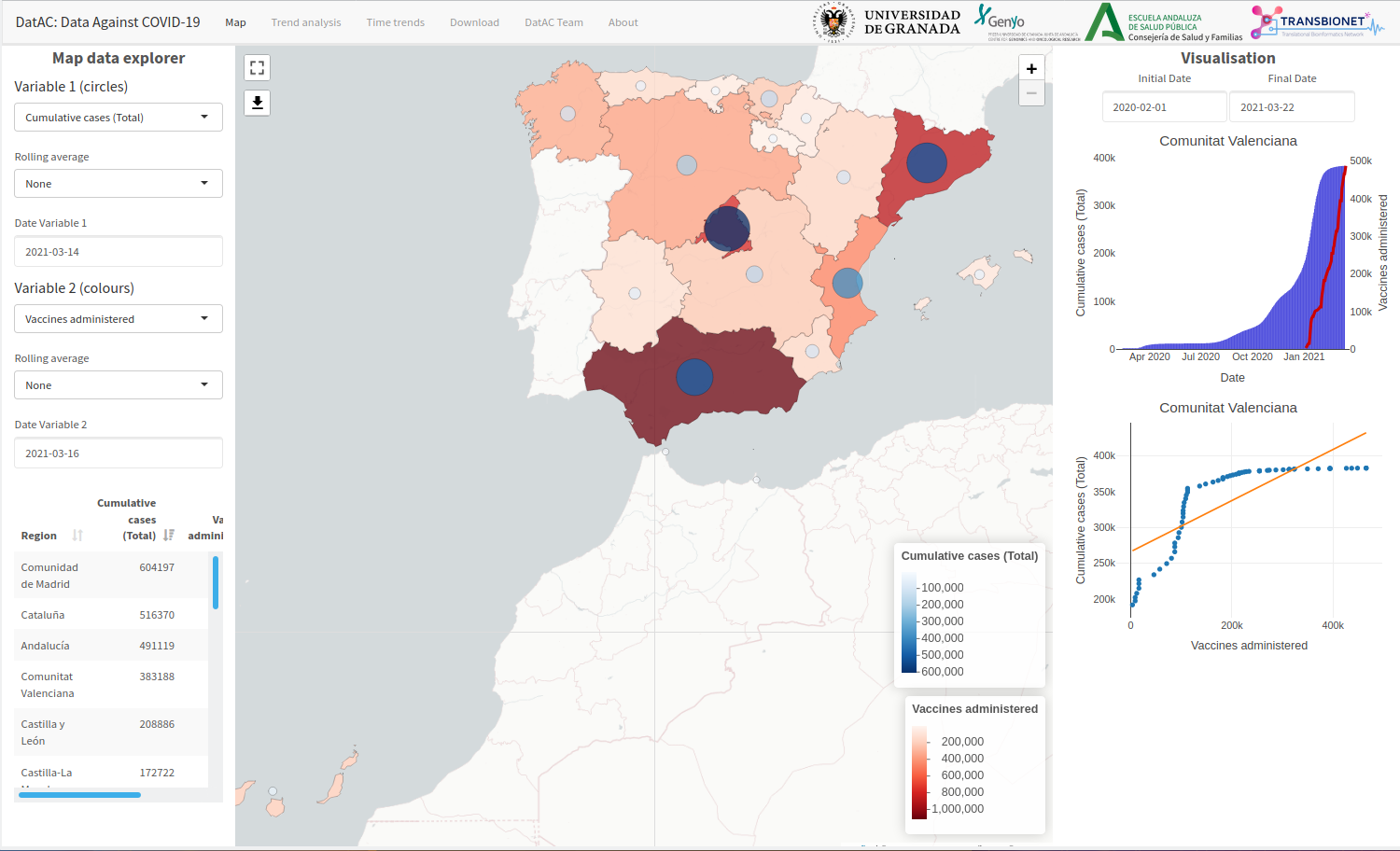
Data Against COVID-19 (DatAC)
Developed by Bioinformatics Unit, GENyO and Statistics Department, University of Granada
Platform that integrates information from COVID-19 with information on environmental and meteorological factors with temporal space aggregation by Spanish provinces and autonomous communities. Not only does DatAC centralize and integrate this type of data, but also implements different analysis and visual exploration functionalities that allow researchers to analyse and look for patterns among the different data sources.

COVID-19 DisGeNET data collection
Developed by Integrative Biomedical Informatics, GRIB (IMIM-UPF) and MedBioinformatics Solutions
The text mining pipeline used in DisGeNET has been customized to scan the literature and identify any mention of genes, diseases and phenotypes, together with mentions of the COVID-19 virus. These mentions are normalized to standard vocabularies. This information is being updated regularly and is publicly available. DisGeNET is an ELIXIR Recommended Interoperability Resource and part of the ELIXIR COVID-19 resources.
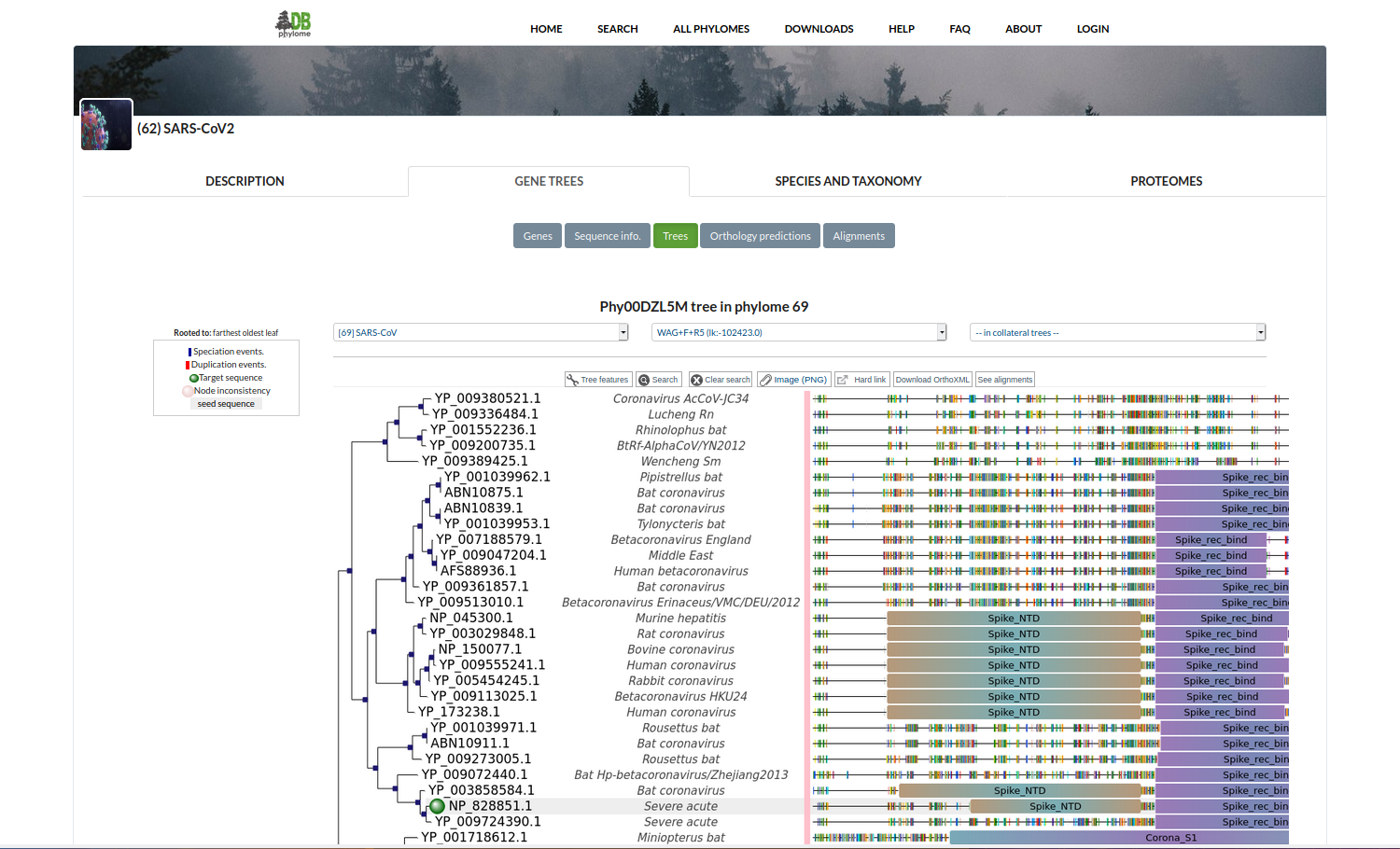
Coronavirus Phylomes
Developed by Comparative Genomics group, BSC and IRB Barcelona
PhylomeDB includes a full phylogenomic analysis of 60 genomes belonging to the Coronavirinae suborder. Three phylomes have so far been built for this collection: one for each of the three main viruses causing human illnesses that recently shifted host from other animal species: SARS-CoV-2, SARS-CoV and MERS-CoV. Gene phylogenetic trees and multiple sequence alignments can be browsed and downloaded.
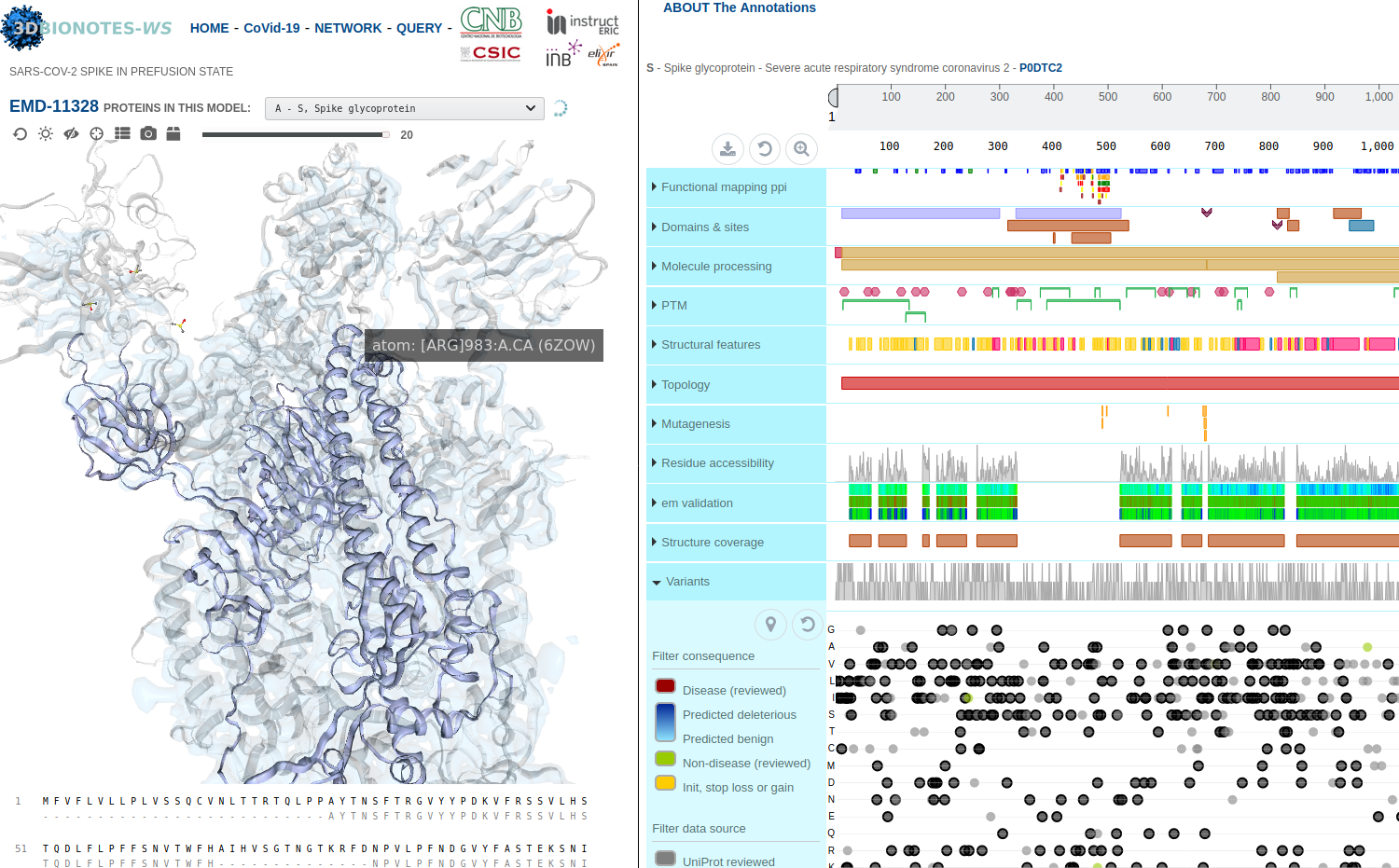
3DBIONOTES-COVID19 tool
Developed by Biocomputing unit, CNB
The new 3DBIONOTES-COVID19 tool enables the study of antiviral targets and the visualisation and annotation of SARS-CoV-2 protein structures. The tool contains SARS-CoV-2 protein models from numerous sources: PDB-Redo, SwissProt, AlphaFold, European Projects and the repository of the Coronavirus Structural Taskforce. This resource is part of the 3DBIONOTES-WS, which its API is an ELIXIR Recommended Interoperability Resource and part of the ELIXIR COVID-19 resources.
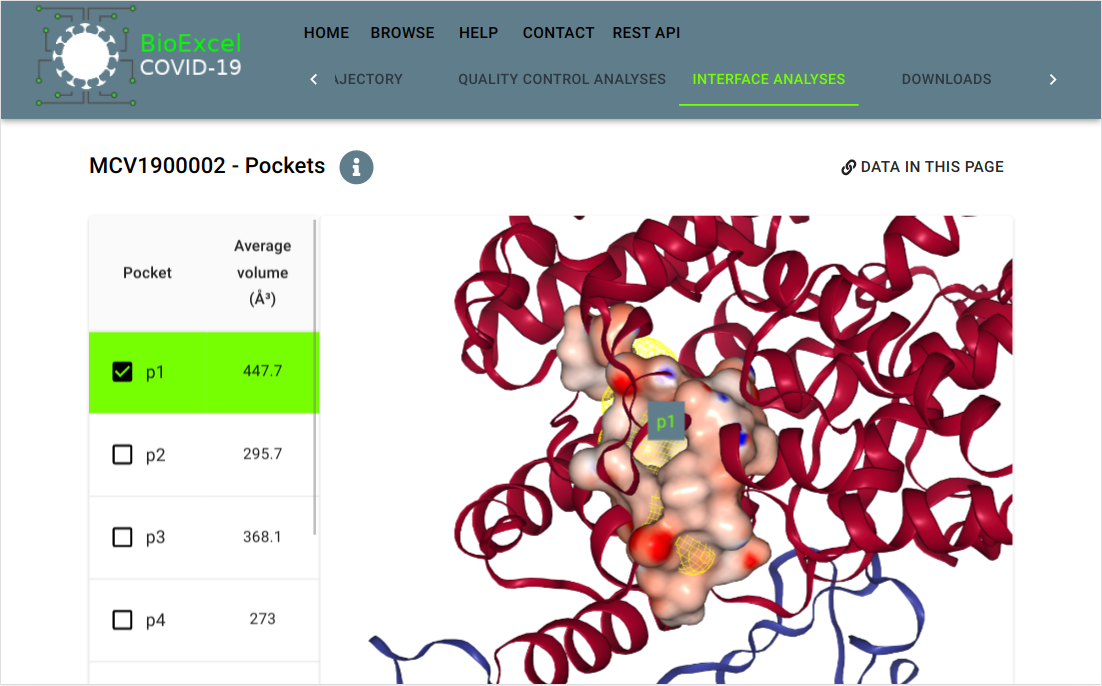
BioExcel-Covid19 (BioExcel-CV19)
Developed by Molecular Modeling and Bioinformatics unit, IRB Barcelona and INB/ELIXIR-ES Computational node, BSC. Resource developed as part of the BioExcel CoE.
Platform designed to provide web-access to atomistic-MD trajectories for macromolecules involved in the COVID-19 disease. BioExcel-CV19 web server interface presents the resulting trajectories, with a set of quality control analyses and system information. All data produced by the project is available to download from an associated programmatic access API.

EPIDEMIXS Coronavirus
Developed by Integrative Biomedical Informatics, IMIM-UPF, as content contributors and advisors
EPIDEMIXS Coronavirus: web app that makes available to the community (general public and professionals) trustworthy sources of information related to the COVID-19 at the national level: studies, articles, protocols, videos.

viralrecon
Developed by Core bioinformatics Unit, ISCIII, and collaboratively updated with the nf-core community.
Bioinformatics analysis pipeline used to perform assembly and intra-host/low-frequency variant calling for viral samples. The pipeline supports short-read Illumina sequencing data from both shotgun and enrichment-based library preparation methods. The pipeline is built using Nextflow, it comes with Docker containers and automated continuous integration tests that run it on a full-sized dataset using AWS cloud.

Collaborative Spanish Variant Server (CSVS)
Developed by Clinical Bioinformatics Area, FPS
A crowdsourcing initiative to provide information about the genomic variability of the Spanish population to the scientific/medical community. It is useful for filtering polymorphisms and local variations in relevant genes for SARS-CoV-2 and in the process of prioritizing candidate disease genes.